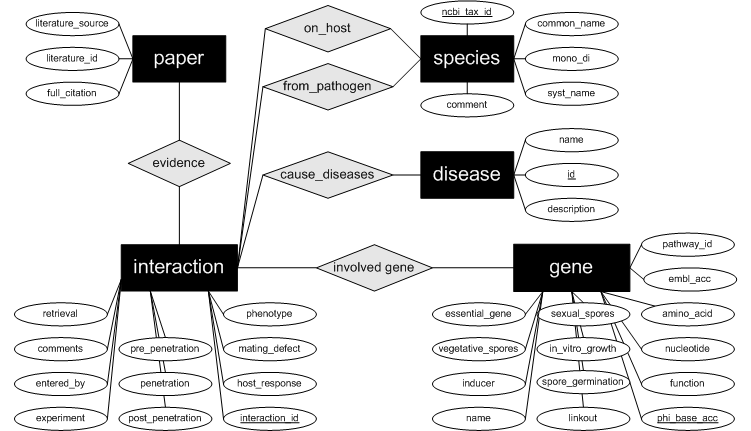
|
Column name
|
Description
|
|
PHI-base
accession
|
Stable
accession number for each database entry to aid curation
|
|
Obsolete accession
|
PHI-base record was replaced with new version or deleted because of duplication
|
|
EMBL
accession
|
Link-out
to EMBL Nucleotide Sequence Database
|
|
Locus ID
|
Obtained from genomic sequence
|
|
Gene
name
|
Name
of the fungal gene that was disrupted in the published study
|
|
Genome location
|
Chromosome number where gene is located or name of pathogenicity island
|
|
Amino
acid sequence
|
Amino
acid sequence of the gene product
|
|
Nucleotide
sequence
|
Nucleotide
sequence of the gene
|
|
Multiple mutation
|
PHI-base accessions of other genes in case multiple gene mutations were tested in one strain
|
|
Pathogen
NCBI Taxonomy ID
|
NCBI
taxonomy ID of the pathogenic species
|
|
Pathogen
species
|
Systematic
name of the pathogenic species
|
|
Strain
|
Strain, in which gene function was tested
|
|
Disease
name
|
Name
of the disease caused by the pathogen host interaction
|
|
Monocot
/ Dicot plant
|
Number
of cotyledons, if the host is a plant
|
|
Host
NCBI Taxonomy ID
|
NCBI
taxonomy ID of the host organism
|
|
Experimental
host
|
Common
name of the host organism
|
|
GO annotation
|
Gene ontology annotation that describes gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent manner (http://www.geneontology.org).
|
|
Pathway
|
Name
of the pathway the disrupted gene is involved in
|
|
Phenotype of mutant
|
Definition of phenotypes:
- Loss of pathogenicity - the transgenic strain fails to cause disease
- Reduced virulence - the transgenic strain still causes some disease formation but this is less than the wild-type strain (ie. a quantitative effect). Synonymous with the term reduced aggressiveness.
- Unaffected pathogenicity - the transgenic strain which expresses no or reduced levels of a specific gene product(s) has wild-type disease causing ability
- Increased virulence (Hypervirulence) - the transgenic strain causes higher levels of disease than the wild-type strain
- Effector (plant avirulence determinant) - currently a plant pathogen specific term which was previously known as an avirulence gene. An effector gene is required for the direct or indirect recognition of a pathogen only in resistant host genotypes which possess the corresponding disease resistance gene. Positive recognition leads to activation of plant defences and the pathogen fails to cause disease. Note some effector genes are required to cause disease on susceptible hosts but most are not.
- Lethal - the transgenic strain which expresses no or reduced levels of a specific gene product(s) is not viable. The gene product is essential for life.
- Enhanced antagonism - the transgenetic strain of a endophyte which shows no asymptomatic colonisation, but is gaining the upperhand of the pathogen-host-interaction.
- Wild-type mutualism - the transgenetic strain of a endophyte which shows no difference in the pathogen-host-interaction comparing to the wild-type
|
|
Mating
defect
|
Yes/No
If the gene disruption causes a mating defect affecting pathogenicity
|
|
Pre-penetration
defect
|
Yes/No
If the gene disruption causes a block in the disease process before
penetration e.g. Formation of appressoria
|
|
Penetration
defect
|
Yes/No
If the gene disruption causes a block in the disease process at penetration
|
|
Post-penetration defect
|
Yes/No
If the gene disruption causes a block in the disease process after
penetration
|
|
Vegetative
spores
|
Defects
in asexual sporulation caused by the gene disruption e.g reduced sporulation
|
|
Sexual
spores
|
Defects
in sexual sporulation caused by the gene disruption e.g reduced sporulation
|
|
In
vitro growth
|
Growth
defects in culture caused by the gene disruption e.g. reduced growth
|
|
Spore
germination
|
Defects
in spore germination caused by the gene disruption
|
|
Essential
gene
|
Lethal
effect from gene disruption
|
|
Inducer
|
For
cases where a particular compound is needed to induce gene expression e.g.
Pectin
|
|
Host
response
|
Details
any difference in the host defence response to a pathogen with a disrupted gene
|
|
Experimental
evidence
|
-
Gene disruption
-
Gene disruption; gene mutation
-
Gene disruption; gene mutation; characterised
-
Gene disruption; gene deletion; complementation
-
Gene disruption; gene deletion
-
Gene disruption; complementation
-
Gene disruption; complementation; other evidence
-
Gene disruption; altered gene expression / gene regulation: overexpression
-
Gene disruption; altered gene expression / gene regulation: overexpression; complementation
-
Gene deletion
-
Gene deletion; altered gene expression / gene regulation: overexpression
-
Gene deletion; complementation
-
Gene deletion; complementation; biochemical evidence
-
Cluster gene deletion
-
Altered gene expression / gene regulation
-
Altered gene expression / gene regulation: downregulation
-
Altered gene expression / gene regulation: overexpression
-
Altered gene expression / gene regulation: silencing
-
Altered gene expression / gene regulation: down- and upregulation
-
Altered gene expression / gene regulation; complementation
-
Biochemical analysis
-
Biochemical analysis; mutation: characterised
-
Biochemical analysis; mutation: characterised; complementation
-
Functional test in host
-
Functional test in host: direct injection
-
Functional test in host: transient expression
-
Mutation
-
Mutation: characterised
-
Mutation: characterised; complementation
-
Complementation
-
Sequence analysis of sensitive and resistant strains
-
Sexual cross, sequencing of resistance conferring allele
-
Other evidence
|
|
Species Expert
|
Research scientist outside Rothamsted with excellent knowledge on specific pathogen species, who contributes to databaseby suggesting new entries and by revising existing entries
|
|
Entered
by
|
Name
of the curator who entered the interaction to the database
|
|
Reference
|
Related literature to the database entry, linked to PubMed or via DOI to the full publication where possible; Fulltext reference is given, if the article is not available online.
|
|
Comments
|
Field
provided for any further free text information
|
|
CAS
|
CAS Registry Number (often referred to as a CAS Number)
|
|
Anti-infective (Chemical)
|
Chemical name
|
|
Compound
|
Accepted (or proposed) common name for an individual active ingredient expected to appear on the product label as definition of the product.
|
|
Target site
|
Target Site of Action: The biochemical mode of action is given.
|
|
Group name
|
The Group names listed are widely accepted in literature. They are based on different sources (mode of action, first important representative, chemical group).
|
|
Chemical group
|
Sub-grouping due to chemical considerations.
|
|
Mode in planta
|
Describes how chemical protects plant against pathogen attack, i. e.systemic - taken up by the plant's leaves or roots and spread throughout the plant's system. |
|
FRAC CODE
|
Numbers and letters are used to distinguish the anti-infective according to their cross resistance behaviour (see www.frac.info).
|
|
Comments
|
Field provided for any further free text information for anti-infective information
|